Their
goal was to help develop a new method for scientists to examine how damage to
our DNA can lead to cancer, and how our cell’s own error correction systems
could offer new treatments.
DNA Error Correction and Cancer
The DNA in our cells can easily become damaged; whether through errors that creep in during replication, exposure to ionising radiation, genotoxic drugs, or even harm caused by pathogens. Fortunately, for relatively minor damage, repair pathways exist which allow a cell to 'reconstruct' the damaged DNA. We can imagine the repair process to be rather like how, if a few l_tt_rs are m_ssed out of a w_rd, our brains fill in the gaps because we know what should be there. If the damage is too severe to repair, the cell undergoes apoptosis; effectively, it self-destructs.
This self-correcting system reduces the likelihood of error-ridden DNA being passed on to the next generation of cells – or, ultimately, the next generation of human. However, not every form of DNA damage can be repaired, and not every repair which takes place is perfect. When errors are not corrected, and propagate to daughter cells, this can be a cause of cancer – the unchecked growth of genetically abnormal cells.
One particularly severe type of DNA damage is a Double Stranded Break (DSB). As the name implies, DSB's are where both strands of the DNA double-helix are severed. Even the tiniest error in correcting a DSB may lead to insertions, translocations, deletions, and chromosome fusions which could leave the genome even more unstable than it was due to the original DSB.
Which of the several repair pathways available is used to (attempt to) repair a particular DSB depends on several factors. The exact nature of the break, the local chromatin context (chromatin is the complex of DNA and protein which makes up the double-helix), and what stage the cell is at in its cell cycle are all known to influence which repair pathway is used.
Understanding the mechanisms which decide the 'choice' of repair pathway is an important step in creating targeted interventions to kill cancer cells. There are already cancer treatments which exploit the apoptotic response, causing damage to the DNA of the cancer cells which is so severe as to cause cell death (for example, PARP inhibitors in Brca-1 deficient cancers). A better understanding of the processes behind DSB repair, and apoptosis, could lead to similar treatments for other cancers.
Creating the right kind of DNA damage
To study the response to DSBs, researchers need first to induce damage to DNA which they can study. Up to now, that damage has generally been inflicted using irradiation, or genotoxic drugs. While irradiation and genotoxic drugs allow different cell types to be studied, the damage they cause is untargeted and not useful for understanding DSB response in context.
More recently, techniques have been developed using the inducible expression of restriction endonucleases – enzymes which sever the DNA double helix at specific locations, on demand. Using these enzymes has allowed for DSB repair pathways to be studied in specific genomic contexts. However, because each endonuclease is specific to a particular cut site, the range of chromosome regions which can be studied is limited.
To tackle this problem, researchers from the Universities of Kent and Sheffield, and UCL, worked with CLF's Octopus team to develop a methodology to induce a tuneable quantity of breaks across a wide range of genomic contexts. To do this, they used a programmable endonuclease (CRISPR-Cas9) combined with a synthetic promiscuous RNA guide. Rather than the usual specific guide, which is designed to target a cut to a specific site to allow for gene insertion, the promiscuous guide makes cuts in the DNA at several points. By choosing a guide which matches a DNA sequence which appears, e.g., 50, 100, or 150 times, it's possible to choose how many (roughly) breaks in the DNA are created.
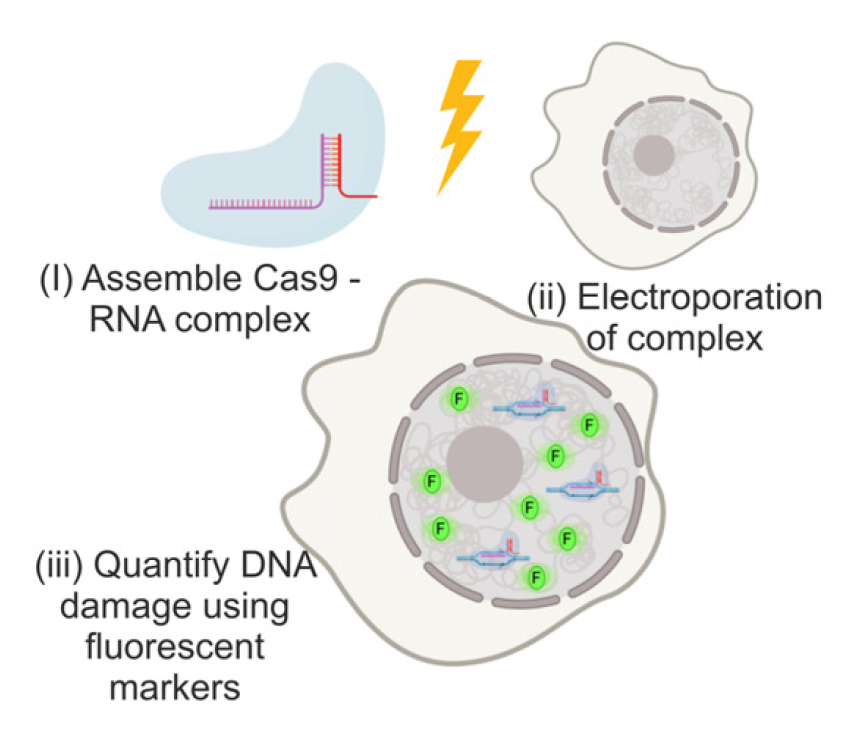
A cartoon depicting the methodology. (i) Recombinant Cas9 RNA guide. (ii) The complex is then electroporated into the mammalian cell line. (iii) Fluorescent markers are used to quantify the level of DNA damage.
Using a fluorescent stain to highlight the sites where DNA had been damaged, and the CLF's Octopus for super-resolution imaging, the team were able to view both 2D and 3D maps of the precise locations of DNA damage over time, as well as measuring the average fluorescence of each cell. There was a good correlation between the expected number of cuts and the observed number of fluorescing foci / mean brightness, proving the new technique can induce a predictable number of DSBs across a wide range of genomic contexts.
Lin Wang, one of the Octopus team, described the super resolution microscopy done on Octopus as 'crucial' to understanding the molecular distribution of the DNA damage.
This new method offers researchers a versatile, fast, low-cost, titratable method for inducing DSBs in any electroporatable cell line, allowing for a systematic exploration of DSB repair in a range of cell and tissue types. It may be that future research leads even to an 'atlas' for each cell type, detailing which DNA repair pathways are invoked in different genomic contexts, and how this relates to the number of breaks in the cell.
Read the paper, published in biomolecules, at https://doi.org/10.3390/biom11020288.